SddCharacters
DonovanSharp - Thu Sep 07 2006 - Version 1.17
Parent topic: SddContents
SDD Part 0: Introduction and Primer to the SDD Standard
3.6 Characters
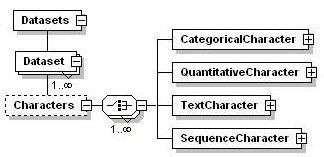
Characters and their states are fundamental elements used in SDD to describe a taxon, specimen or other entity. Characters and states (and their arrangement into a hierarchy) provide the ontology for the SDD document. Sdd employs a single flat character list containing states. The order of characters in SDD is not informative and is instead defined exclusively in the <CharacterHierarchies> element. This enables different character sequences for different reporting purposes and splitting characters into multiple files or reusing characters from different terminologies.
SDD States may be defined either locally within a character or enabled by reference to ConceptStates.
SDD supports four character types, dealt with individually below.
3.6.1 Categorical characters
Categorical characters express either naturally discontinuous states or categories defining parts of a continuous range. Examples include presence (present/absent), colour (red/blue) and shape (round/ovate).
A simple SDD code instance representing a categorical character definition has the basic structure shown below, Example 3.6.1.1 shows two categorical characters as traditionally expressed; Example 3.6.1.2 shows the same characters and states represented in SDD.
Example 3.6.1.1 - Traditional representation of categorical characters.
|
Example 3.6.1.2 - SDD representation of the categorical characters in 3.6.1.1.
|
Characters and their states are represented using a label, perhaps in a defined language and for a defined audience. States for a categorical character are listed under their appropriate character. Elsewhere in SDD documents, characters and states are referred to by their id values rather than by their labels.
Note that characters and states defined in the <Characters> element form a flat list. The <CharacterHierarchies> element is used to arrange characters defined here into a hierarchy.
Note that a <CategoricalCharacter> element must have one or more state elements
<Assumptions> allows properties of the character (such as whether the states are ordered or unordered and whether the states are naturally discrete) to be set
<Mapping> allows characters and states to be mapped to each other (e.g. the states narrowly ovate and broadly ovate may be mapped as substates of the state ovate)
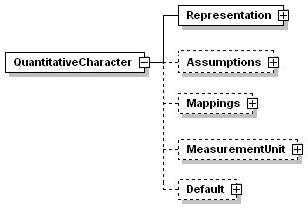
3.6.2 Quantitative characters
Quantitative states record an actual measurement (e.g. number of legs = 8; leaf length = 6.8 cm), range of measurements (e.g. leaf length4.5-10.6), or statistical parameter for a set of measurements (e.g.). Extended ranges (e.g. wing length (5-)10-40(-50) mm, interpreted as wings usually between 10 and 40mm but occasionally down to 5mm or up to 50mm) are supported.
A simple SDD code instance representing a quantitative character definition has the basic structure shown below and in Example 3.6.2.1.
Example 3.6.2.1 - SDD definition of a simple quantitative character “Leaf length”.
|
<Assumptions> allows properties of the character to be set. Properties include whether the values are expected to be integers, discrete or continuous etc, and whether there is an expected plausible range). Any measure used in a description constitutes valid information. However, a list of recommended measures for sets of characters may be defined in concept nodes. <Mapping> allows numeric states to be mapped to categorical character states (e.g. the range 0-10 may be mapped to small, and the range 10-20 to medium).
<MeasurementUnit> defines a measurement unit (mm, inch, kg, °C, m/s, etc.) or dimensionless scaling factor (such as '%') applying to all values of this character. If a Default MeasurementUnitPrefix is defined (see below), this must be entered without a prefix (e. g., 'm' instead of 'mm'). (Measurement units apply only to values plus those statistical measures not marked as IsDimensionless='true'.).
<Default> provides a default value to be used for the character if no value is specifically recorded in a description.
3.6.3 Text characters
Text characters record information not easily ascribed to a measurement or atomised into characters and states. Examples include place of publication (e.g. “Smith 1998. Flora of Erehwon, Z-Publ.”), derivations (e.g. “acuta- from the Latin acuo (sharpen), alluding to the sharply pointed glumes (Aristida acuta)”) or general notes (e.g. “One record for tropical Queensland but mainly recorded from subtropical coastal Queensland to northern New South Wales. Eucalyptus woodlands and forests on poor soil”).
The content of quantitative characters simply exists as plain text within a <Representation> element as in Example 3.6.2.1 below.
3.6.3.1 Sdd representation of text characters
|
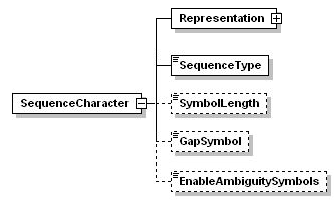
3.6.4 Sequence characters
Sequence characters record the coding of genes or proteins for use in molecular analysis. The basic structure od SDD code for sequence characters has the basic structure shown below and in example 3.6.4.1.
3.6.3.1 Sdd representation of sequence characters
|
<SequenceType> is currently limited to 'Nucleotide' and 'Protein', but future SDD versions may expand this after appropriate discussion. The special nucleotide type RNA/DNA are currently not considered necessary. The symbols U (RNA) and T (DNA) should be considered equal for the purpose of analysis.
<SymbolLength> refers to the number of letters in each symbol. Nucleotides are always codes with 1-letter symbols, but proteins may use 1 or 3-letter codes (A or Ala for alanine). In NEXUS SymbolLength is implicit in the Token command.
<GapSymbol> is a string identifying the 'gap' symbol used in aligned sequences. The gap symbol must always be SymbolLength long. A gap is a place where no data exist, but where a position must be filled because it is assumed that sequence symbols were inserted or deleted during evolution.
<EnableAmbiguitySymbols> provides support for ambiguity symbols such as R, Y, S, W for nucleotides, or B,Z for proteins in the sequence string.
-- Main.DonovanSharp - 01 Jun 2006